
Structural biology
The Swedish NMR centre is an international, national and local research infrastructure that provides access to state-of-the-art NMR instrumentation for structural biology projects.
We offer access to specialist support and NMR spectrometers for solution NMR in the range between 700 MHz and 900 MHz. Our spectrometers are equipped with state-of-the-art probes for unsurpassed sensitivity, and have contributed to publications in multiple areas of research. The Swedish NMR centre comprise two nodes, one in Gothenburg and one in Umeå, together encompassing a facility within SciLifeLab.
Within SciLifeLab, we are coordinating the platform Integrated Structural Biology, acting as a spider in the web for you to get in contact with infrastructures offering the techniques you need in your project. A successful protein project starts with protein expression (PPS), goes through biophysical characterisation and sample optimisation for the technique in focus (ProLinC), continues with atom resolution methods (structural proteomics, NMR, Cryo-EM, x-ray crystallography) and data can be analysed in combination with computational structural biology, i.e. AlphaFold (NBIS).
Links to relevant information
Isotopic labelling of proteins for NMR purposes
The NMR active nuclei naturally occurring in proteins are 1H, 13C and 15N. 1H is naturally occurring at >99%, while 13C and 15N needs to be enriched by recombinant expression of the protein in a cell medium containing these isotopes. Different labelling schemes are depicted below and what possible NMR experiment they are associated to. 19F can be introduced at specific sites, and this can be done either in vivo or in vitro using the SNC in-house cell-free system. You can also find approximate costs for 1L of expression medium.
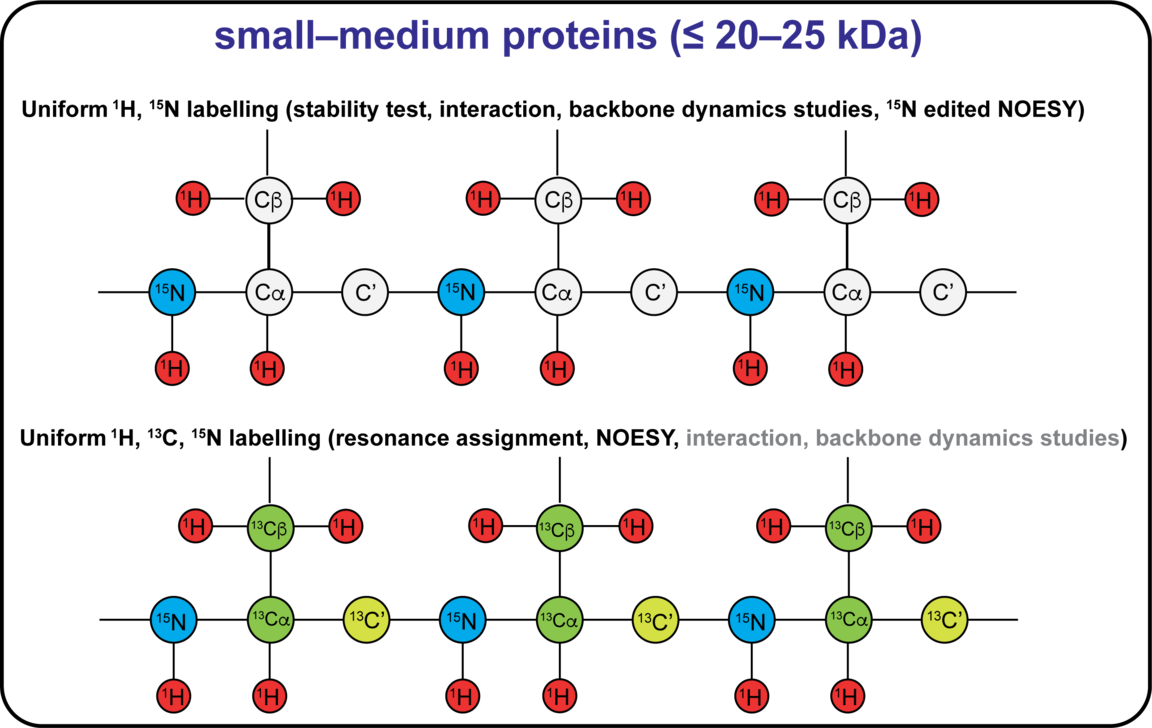
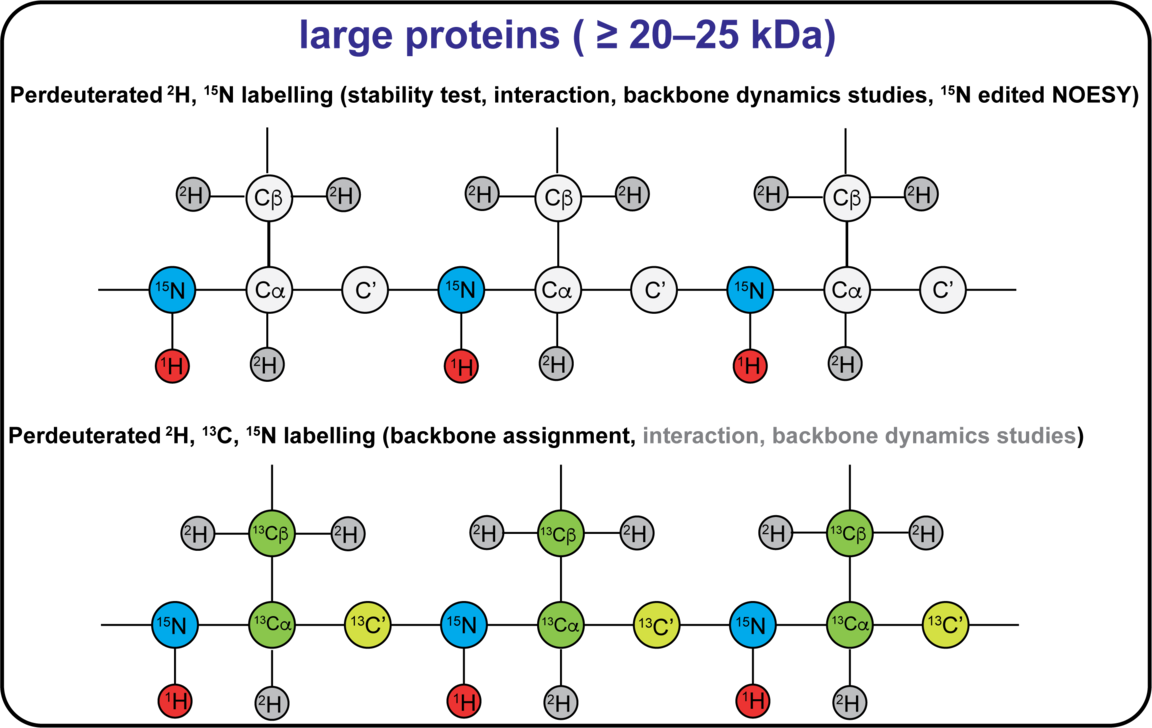
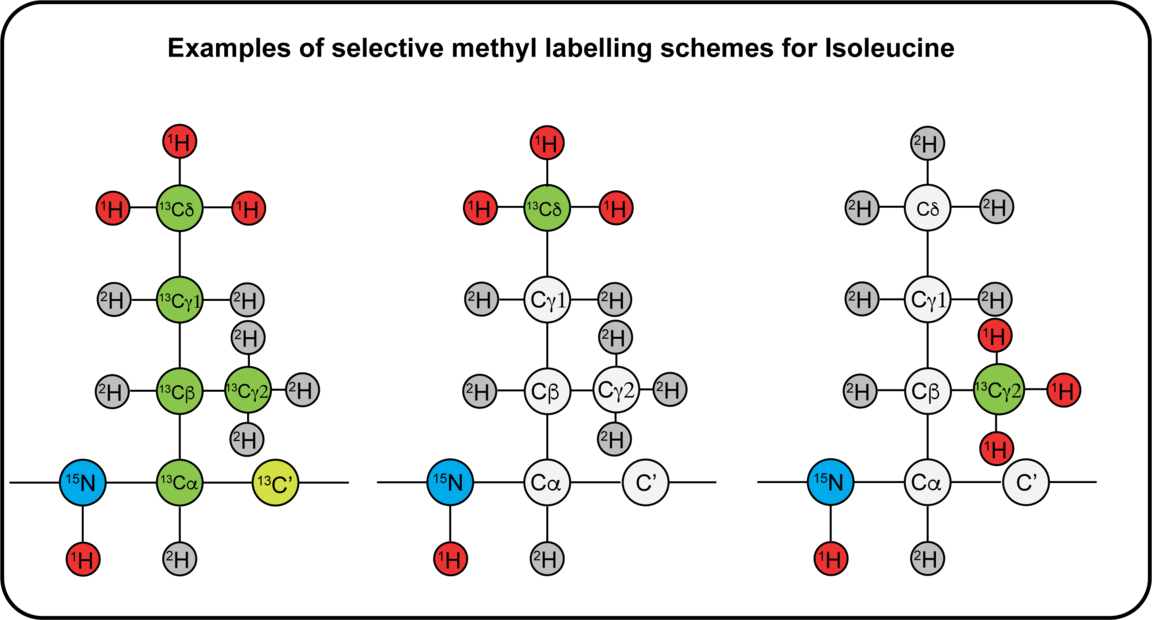
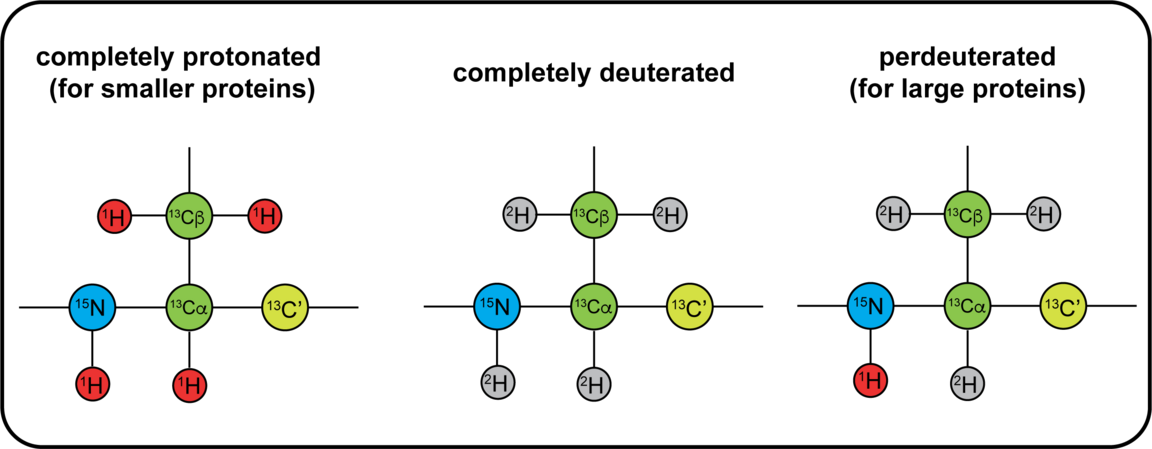
Sample requirements for NMR
Ideally, samples for structural biology studies should not contain proton-rich buffers or additives. If that can not be avoided, a deuterated version of the buffer substance/additive is strongly recommended for most kinds of measurements. The pH of the sample should be kept below 7 to reduce the exchange of the labile amide protons with bulk water, which otherwise leads to a decrease in signal intensity. For more details on sample requirements, see drop-down menus below for the different uses of NMR in structural biology.