
Marine Genetic Models
In the project IMAGO, Infrastructure for MArine Genetic model Organisms, we develop marine species into new experimentally amenable genetic models. IMAGO provides reference genomes and additional genomic resources, for example, gene maps and transcriptomes, to interested collaborators. Also, we are happy to share knowledge about how to culture the model species. Further development is ongoing with improved annotations and new versions of reference genomes for some species.

The IMAGO species represent key-species in North Easts Atlantic coastal ecosystems. Check details for each species below, and get in contact with us for more information.
The species
Brittle star, Amphiura filiformis
Bay barnacle, Balanus improvisus
Marine yeast, Debaryomyces hansenii
Bladderwrack, Fucus vesiculosus
Baltic isopod, Idotea balthica
Rough periwinkle, Littorina saxatilis
Sand goby, Pomatoschistus minutus
Marine diatom, Skeletonema marinoi
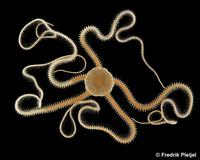
Brittle star, Amphiura filiformis
Amphiura filiformis is a burrowing brittle star with a disc up to 10 mm in diameter with long arms (10 x disc diameter in average) that extend into the water column for suspension feeding, making it an ecosystem engineer and important food item for flatfish, cod and crayfish. It is the dominant species on many sublittoral soft bottoms down to 200 m depth in the North Sea and the Mediterranean. Amphiura filiformis has exceptional adult regenerative abilities and can regenerate fully functional arms in a matter of weeks. A. filiformis has separate sexes and reproduce annually during the summer months (July-September). Adults are at least 3 years old before they are mature and have a life span of 10-20 years. Larvae is planktotrophic and metamorphose a month later under culture conditions. Both larvae and adults can be cultured in our facilities.
Contact
Bay barnacle, Balanus improvisus
The Bay barnacle is a sessile crustacean found in coastal, shallow waters worldwide, especially in estuarine and brackish waters. Being sessile, barnacles experience extreme changes in environmental conditions requiring specific evolutionary adaptations. It is one of few macro-invertebrates that inhabits most of the salinity gradient from the North Sea to the Baltic Sea and is known for its broad salinity tolerance (1.8 – 35‰) The species has a complex life cycle with seven different larval stages, which have been studied both in ecological contexts as well as in antifouling research.

Contact
- Anders Blomberg
- Magnus Alm Rosenblad (bioinformatician)
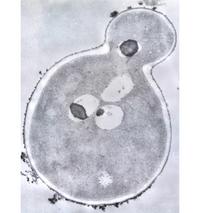
Marine yeast, Debaryomyces hansenii
Debaryomyces hansenii is a marine yeast that occurs globally with extreme tolerance to salt/dehydration stress. It is a unicellular free-living organism that mostly occurs in an asexually reproducing haploid state, and it is slightly salt-stimulated and grows best under salt levels in the range of sea-water. Its adaptation to salt also makes it frequently isolated from salt-conserved foods.
Contact
- Anders Blomberg
- Tomas Larsson (bioinformatician)
Bladder wrack, Fucus vesiculosus
Bladder wrack is a foundation species in the North Atlantic and is essentially the only perennial species of macroalga present in the northern parts of the Baltic Sea. It provides important habitat for associated flora and fauna, contributing to maintain biodiversity in shallow rocky areas. Bladder wrack reproduces mainly sexually but is also reproducing asexually by fragmentation in the northern and eastern parts of the Baltic Sea.

Contact
Baltic isopod, Idotea balthica
The isopod Idotea balthica is an ecosystem engineer which increases productivity of seagrass beds and seaweed forests by reducing epiphytes. I. balthica is a major part of the diet of many fish species, including cod and, in the Baltic Sea, perch. The species is found both sides the northern Atlantic. It is very common in the Baltic Sea where it lives in association with seagrass and fucoid seaweeds down to salinities of 4 psu. I. balthica has dimorphic sexes and the brooded offspring develop without planktonic stages.

Contact
Rough periwinkle, Littorina saxatilis
The rough periwinkle Littorina saxatilis is a dominant grazer on filamentous and microalgae on rocky shores. It has a wide distribution in the North Atlantic from Greenland to Portugal, and from Baffin Island to Delaware Bay. It commonly reaches densities of several hundred snails per square meter and its removal completely changes the ecosystem of rocky shores. It is an important food source for crabs, and in some areas birds and fishes. Unlike many marine invertebrates, L. saxatilis lacks larval stages and females give birth to small shelled juveniles. This greatly limits dispersal of the snails; however, occasional long-range dispersal occurs by rafting. The species exhibits an exceptional morphological variation, which confused taxonomists for centuries, and received the title of the “Champion of taxonomic redundancy” from the World Register of Marine Species.

Contact
Sand goby, Pomatoschistus minutus
The Sand goby inhabits shores of the North East Atlantic and the Baltic Sea. Its behaviours and reproductive strategies include exclusive paternal care with males building nests under mussel shells. The Sand goby is euryhaline, i.e. it is able to adapt to a wide range of salinities, however, not to fresh water. Sand goby life stages from pelagic fry to adults are staple food for cod and other large commercial fishes.

Contact
Marine diatom, Skeletonema marinoi
The chain-forming marine species Skeletonema marinoi is an important primary producer in the North Atlantic. It is especially abundant during spring bloom, when it reaches densities of millions of cells per litre. The predominant means of propagation is through vegetative division, but sexual reproduction occur and resting spores are formed under some conditions .
