Computational and theoretical biochemistry
Short description
Leif Eriksson's research group uses advanced computer simulations to develop new drug molecules or describe the structure, function and interactions of proteins. To aid in their work, they rely heavily on the use of Swedish supercomputing clusters. Their research focuses to a large degree on the identification of new compounds for treating cancer, for example by blocking the cells abilities to protect themselves against stress. The group also develops models of protein complexes and study how the cells read DNA (so-called transcription).
Our research
In our research, we use advanced computational chemistry and bioinformatics based tools such as homology modeling, protein-protein docking, virtual screening, molecular dynamics simulations and QM/MM methods, to develop new compounds that can be used as drug molecules, or to better understand the functions and interactions of proteins.
A large part of our work is directed towards cancer therapy, where we study a range of potential targets. In recent years, we have focused on a protein called IRE1 (inositol requiring enzyme 1) that is involved in the cellular response to / protection against stress, the unfolded protein response (UPR). Amongst other things, we explore how different proteins come together in a dynamic complex with IRE1, the ‘UPRosome’, and we have developed the first BBB penetrating inhibitor targeting the IRE1 kinase site. This compound has with great success been tested in preclinical experiments for treatment of the lethal and aggressive brain tumor glioblastoma multiforme (GBM).
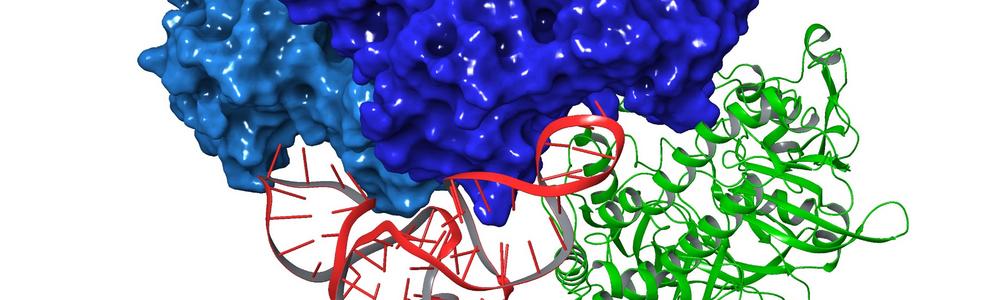
Besides IRE1 and several other potential targets for cancer therapeutics, we also work with new methodology to better predict the structures of protein complexes, and machine learning in drug design. A part of our team is also conducting research on transcription factors and how these are able to recognize the ‘right’ sequence in DNA, and how DNA flexibility is governed by the specific order between nucleic acids.
Artificial Intelligence and Machine Learning
Our research group has thus far developed two software tools, based on advanced machine learning methodology.
The code i-TripleD is able to determine dissociation constants of ligands with extreme speed and accuracy. Recently, we screened a database of 1 billion compounds in less than 36 hours; a process that with conventional docking techniques would require five months of supercomputing time.
Our second invention is the programme iPIN. With this programme, we can with close to 100% accuracy determine which proteins that will interact.
Both these methods are currently subject to patenting, and has in a collaboration with Chalmers Ventures and Chalmers School of Entrepreneurship led to the formation of the spin-out company ANYO Labs. Further work in the area of ML-based methodology is currently in the pipeline.


