
Joan Camuñas-Soler Group
Short description
Joan Camuñas-Soler, PhD, Assistant Professor in Bioinformatics for Translational Medicine.

Dr. Camuñas-Soler joined Sahlgrenska Academy and the Institute of Biomedicine as an Assistant Professor of Bioinformatics for Translational Medicine in 2022.
Camuñas-Soler received postdoctoral training in the lab of Stephen Quake at Stanford University where he developed new tools in single-cell genomics and non-invasive diagnostics to investigate metabolic disorders and pregnancy complications.
In addition to his academic work, at Stanford he was involved in the founding of Mirvie, a molecular diagnostics start-up company based in South San Francisco. Prior to that, he obtained his M.Sc. in Biophysics from the Royal Institute of Technology, a PhD in Physics from the University of Barcelona, and he was a visiting scholar at Carlos Bustamante lab at UC Berkeley.
His work has been published in journals such as Science, Nature, Cell Metabolism, Nature Physics, and PNAS, and it has also been recognized with several awards, patents, and prestigious establishment grants.
The Camuñas-Soler lab stands at the interface of genomics, biophysics, and precision medicine. Our mission is to develop new single-cell technologies and non-invasive molecular diagnostic tools to understand and predict cellular dysfunction in human disease. Our research is highly interdisciplinary in nature and combines approaches from molecular engineering, biophysics, and computational biology.
Single-cell genomics and patch-seq electrophysiology
One pillar of our work is the development of new single-cell methods to study cellular activities and characterize dysfunctional cellular states in disease. We are interested in understanding how cellular heterogeneity arises within cell types, and to what extent the transcriptome of a cell is a faithful representation of its physiologic state. The lab has a focus on metabolic disorders such as diabetes, but we are excited to explore other systems where single-cell genomics and data-driven approaches may uncover unexpected biology.
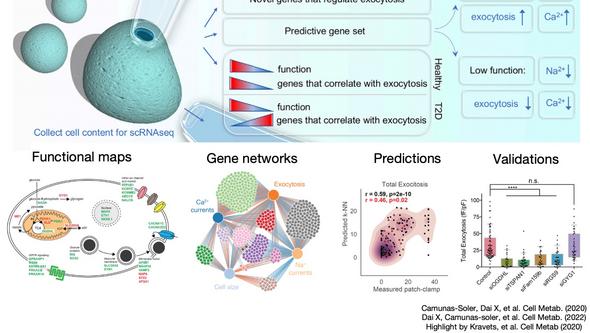
We recently pioneered a method to combine single-cell RNA sequencing and patch-clamp electrophysiology (patch-seq) in islet cells in the pancreas. This is important because key cell types in the islets have been found to exist as different subpopulations; for example, not all β-cells (the source of insulin) are the same in a given pancreas. But how these subpopulations may be functionally different or cause diabetes has been unclear.
Patch-seq has allowed us to characterize the functional response of β-cells and link them to the cell’s transcriptome. In this way, we have identified genes that switch their behavior in diabetes, as well as predicted the electrophysiological properties of a cell based on its gene expression profile. We are currently mapping some of these results to additional islet cell types such as α- and δ-cells. Another major unsolved question is how these findings fit within the complex environment of a tissue or a multicellular structure. For instance, in a pancreatic islet, both (i) cell-to-cell interactions and (ii) electrical coupling have a significant role in insulin secretion. We employ recent experimental advances in genomics, imaging, and electrophysiology to tackle these questions.
From a single-cell genomics perspective, our work is also highlighting that some of the variation or “noise” measured in single-cell experiments can be attributed to variation in cellular features such as exocytosis, cell excitability or simply cell size. To date, only a handful of studies have addressed the challenge of coupling single-cell transcriptomic measurements to additional measures of cell function or general physiology. We are interested in using this type of data to characterize “transcriptional noise” and identify the extent to which it can be traced back to physiological properties of the cell.
Non-invasive diagnostics using cell-free RNA
Another pillar of the group is non-invasive diagnostic measurements using circulating nucleic acids such as cell-free DNA and RNA. Blood circulates throughout the human body and collects molecules from virtually every tissue. Among these molecules, small fragments of nucleic acids circulate in peripheral blood and can be used to develop diagnostic tools by combining next-generation sequencing and advanced computational methods.
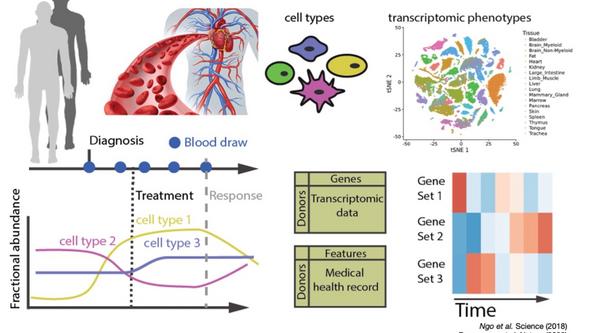
A key feature of cell-free RNA technology is that it enables us to monitor molecular phenotypes by following transcriptomic changes in plasma that reflect changes in cell type composition across the human body. So far, we have shown that this can be used in multiple settings, such as tracking fetal and placental development during pregnancy or to follow immune responses of the host to viral infections.
We are currently developing methods based in the precise quantification of cell-free RNA to investigate tissue disruption and inflammatory responses in multiple human conditions. We are also developing new machine learning algorithms to leverage single-cell RNA sequencing atlases and identify changes in cell type representation during disease onset and as a response to treatment. We envision that these tools will facilitate patient stratification and targeted therapeutics in diseases known to be caused by heterogenous factors. We are continually looking forward to translate our work into clinical use.
We are currently recruiting post-docs, PhD students, and lab technicians.
For more information please contact: joan.camunas@gu.se
We also have openings for MSc thesis projects.

Marta Gironella-Torrent
Postdoctoral Fellow, University of Gothenburg, Sweden.
PhD in Physics, Faculty of Physics, Universitat de Barcelona.
MSc in Advanced Physics, Faculty of Physics, Universitat de Barcelona.
Yike Xie
Postdoctoral Fellow, University of Gothenburg
PhD in Bioinformatics, UNSW Sydney
MSc in Pharmacy, Fudan University


Sandra Postic
Postdoctoral Fellow, University of Gothenburg
PhD in Physiology, Medical University of Vienna
MSc in Molecular Biology, University of Zagreb
Martin Selin
PhD student, Co-supervised (with G. Volpe)
MSc in Complex Adaptative Systems, Chalmers University.


Mercedes Dalman
PhD student
MSc in Engineering Mathematics & Computational Science, Chalmers University
BSc in Biotechnology, Chalmers University
Oliver Forsell
PhD Student
MSc in Biotechnology, Chalmers University
BSc in Biotechnology, Chalmers University


Paolo Valentini
Research Assisstant
MSc in Cell and Molecular Biology, Uppsala University
BSc in Biology, University of Modena and Reggio Emilia
Emma Walsh
Research Assisstant
MSc in Genomics & Systems Biology, University of Gothenburg
BSc in Cellular, Molecular & Microbiology, University of Calgary

Jain, M., Namsaraev, E., Rasmussen, M., Camunas-Soler, J., Siddiqui, F., Reddy, M. Methods And Systems For Determining A Pregnancy-Related State Of A Subject. US Application No.: PCT/US2020/018172
Moufarrej, M.N., Quake, S.R., Melbye, M., Ngo, T.T.M., Camunas-Soler, J. A Noninvasive Molecular Clock for Fetal Development Predicts Gestational Age and Preterm Delivery. U.S. Application No.: 16/758,844 PCT/US2018/057142
Quake, S.R., Camunas-Soler, J. Noninvasive prenatal diagnosis of single-gene disorders using droplet digital PCR. U.S. Application No. 62/530,041 PCT/US2018/041150
Active research grants as Principal Investigator
- Swedish Research Council (VR) (2022-2025) – Establishment grant #2021-05109
- The Wallenberg Centre for Molecular & Translational Medicine (2022-2025) – Starting package & Tenure-track funding
- Swedish Foundations' Starting Grant (2023-2027) - Erling-Perssons stiftelse
Grants and awards by team members
- Marie Curie Postdoctoral Fellowship (2022-2024) - Marta Gironella-Torrent
Institutional Support
- Institute of Medicine, University of Gothenburg (2022-2025)
Awards
- Award AntalGenics SBE-33. Outstanding young biophysicists award of the Spanish Biophysical Society (2018)
- Award XXI Premi Claustre de Doctors Universitat de Barcelona for best PhD dissertations (2017).
- Best Poster Award, 7th International Conference on Unsolved Problems on Noise (2017)
- Travel Award, International StatPhys 25 Conference (IUPAP), Seoul, South Korea. (2013).
- Fellowship Research Stay at UC Berkeley. CIBER Bioengineering Biomaterials and Nanomedicine (2011)
- Fellowship FI from AGAUR, PhD Fellowship (2009)
- Travel Award, NSF-GEM4 Summer School on Cellular and Molecular Mechanics with Focus on Enabling Technologies, University of Illinois Urbana-Champaign, USA. (2009)
- Fellowship APIF, PhD Fellowship, University of Barcelona (2009)
- Fellowship CIBER Lanzadera from CIBER Bioengineering Biomaterials and Nanomedicine (2008)
- Scholarship, Swedish-Spanish Foundation (2006)
Other Memberships
- Co-President of AIMS, Association of Industry-Minded Stanford Professionals, Stanford University (2016-2018).
- Council Member of SURPAS, Stanford Postdoctoral Association. Stanford University (2017).
- Human Islet Research Network (HIRN) (2016-2020).
- Center of Excellence for Stem Cell Genomics (2016-2018), California Institute for Regenerative Medicine.
- Spanish Biophysical Society (2012-2018).
- Co-chair of the Interdisciplinary Meeting of Predoctoral Researchers conference JIPI (2011-2013). University of Barcelona.

