Mapping inflammation and metabolism in chronic disease
Short description
Åsa Torinsson Naluai's research group studies the biology behind chronic inflammatory diseases and searches for biological markers that can be used to identify individuals at risk. Using "-omics" large scale data, we connect molecular pathways to a disease model and have identified new biomarkers for early detection of inflammatory processes. To validate and improve biomarker analysis we collect and develop digital tools using specific sample collection sets for determining sample quality and also using this knowledge in the biomarker discovery data analysis.
Our research goal is to generate new hypotheses on what causes inflammatory chronic disease, and to test these hypotheses in experimental systems. To do this we use “big data” and -omics technologies to map the molecular landscape in healthy individuals versus individuals with a disease. Our research project challenges the view that obesity has a causative role in modern non-communicable diseases like type 2 diabetes and cardiovascular disease. Instead, we believe that specific amino acids cause inflammation and this inflammation leads to both obesity and disease. Below is a short summary of three overlapping but slightly different projects.
Triggers of chronic inflammation
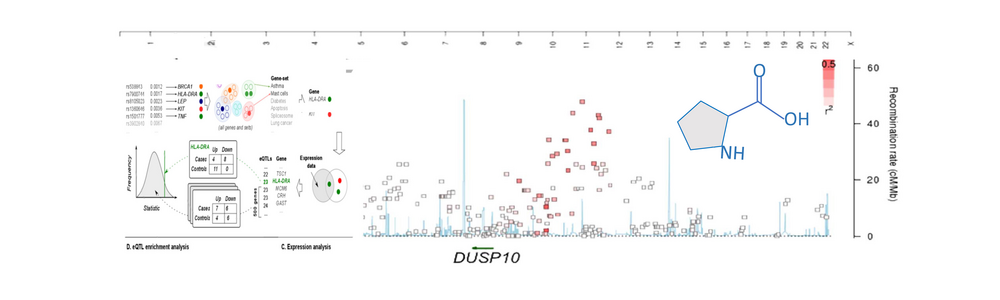
Our strategy has been to utilize bioinformatics tools and study design to combine several advanced molecular technologies (–omics technologies). Our first Genome Wide Association Study (GWAS) was in families with several children affected with celiac disease (gluten intolerance). Together with Staffan Nilsson and Malin Östensson at Chalmers University of Technology, we used novel bioinformatics tools where, for example both epistasis and pathway analyses were combined to get more information from our data. These findings led us to a new hypothesis of why someone might develop chronic inflammation. We currently investigate this hypothesis further using metabolomics (NMR, Mass spectrometry) and transcriptomics (RNA sequencing and qPCR). Patient material that we work with include celiac disease, psoriasis, nasal polyps, recurrent aphthous stomatitis and healthy controls. If our hypothesis is true, molecules signaling nutritional status and specifically amino acids, are in certain settings, part of an ancient signal of “danger”, which the immune system reacts to, and leads to a state of chronic inflammation. In this project we also collaborate with Professor Polly Matzinger at the National Institute of Health in Bethesda USA.
Biobanking and sample quality
To validate and improve -omics analysis, we collect specific biobank sample sets and use machine learning to develop digital tools for determining sample quality. In this effort, we work closely together with scientists at Chalmers University of Technology and at the Universities of Gothenburg and Skövde as well as with advanced technological platforms across Sweden.
COVID-19
During the COVID-19 pandemic, we have been part of initiating a collection of samples with the goal to identify individuals with asymptomatic COVID-19 or “Long-COVID” (those with symptoms not necessarily treated or requiring emergency care). These samples can be used for diagnostic purposes when needed but are also available for various research groups with ethically approved research projects. COVID-19 is tightly linked to diabetes and cardiovascular disease and we plan to investigate the molecular connection between these diseases and to dig deeper in the understanding of how our immunity works.
Åsa Torinsson Naluai
Principal Investigator
Affiliation:
Department of Laboratory Medicine,
Institute of Biomedicine