Super-resolution fluorescence microscopy
Super-resolution in fluorescence microscopy can be defined as all techniques with a resolution higher than the classical diffraction limit of light.
The Centre for Cellular Imaging has an ELYRA 7, which can perform Lattice Structured Illumination Microscopy (SIM) and Single Molecule Localization Microscopy (SMLM). The ELYRA 7 microscope is a widefield inverted microscope designed for high-speed super-resolution imaging of biological specimens with minimal light exposure.
Linear SIM
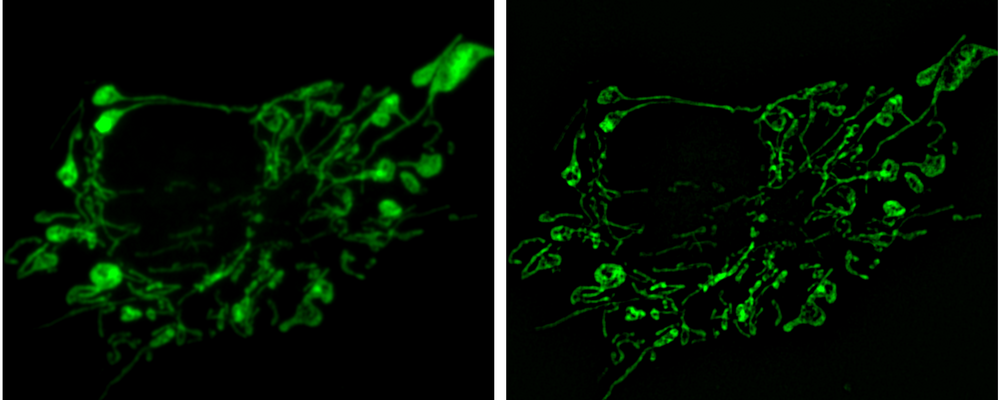
Structured Illumination Microscopy (SIM) is a technique, which overcomes the diffraction limit by using a patterned illumination and hence creating a moaré effect in the image. An image with twice the resolution in all three spatial dimensions can then be calculated from the known illumination pattern and the recorded image.
SIM was pioneered by Gustaffson, and in its first implementation a line grid pattern was used to produce the SIM illumination pattern. This example is shown in the image below [Vangindertael]
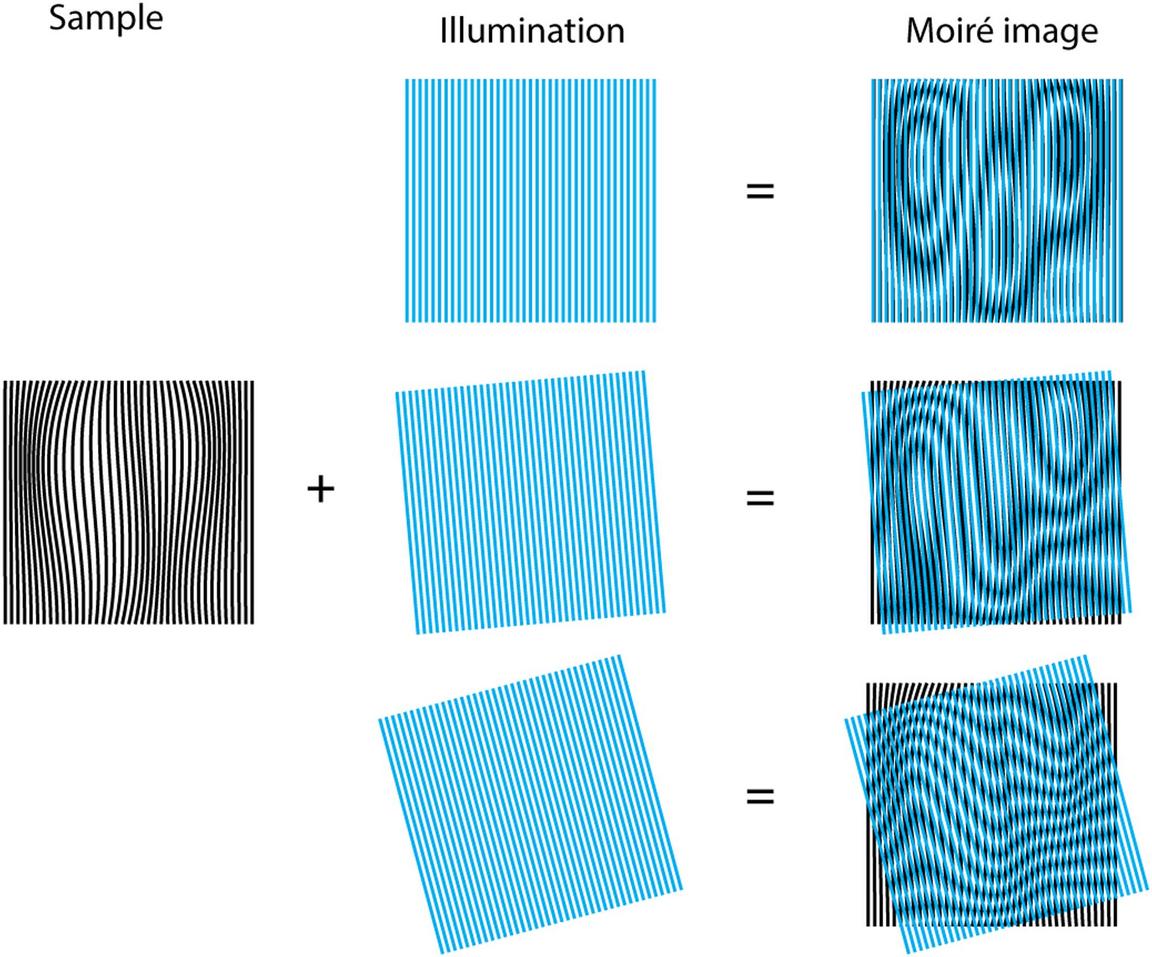
This line structuring element allows to obtain higher resolution along one spatial direction. Therefore, the pattern must be rotated to improve the resolution in other directions and obtain an isotropic image resolution. A common approach was to rotate the pattern on 3 different orientation and translate it on 5 different steps, generating a total of 15 images. These 15 images are then combined to obtain the final super-resolved image. Such rotation is time consuming and thus limits the imaging speed.
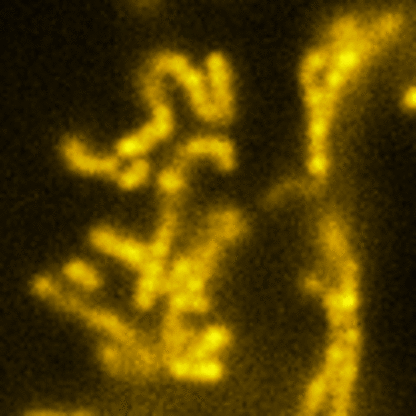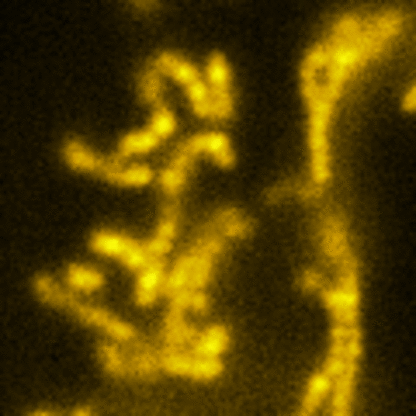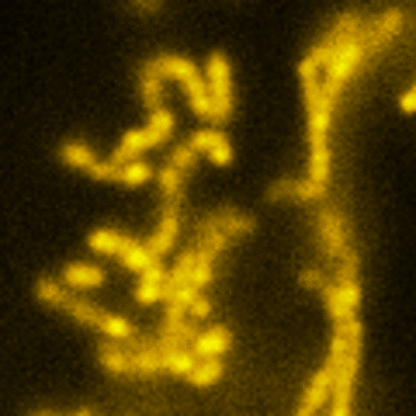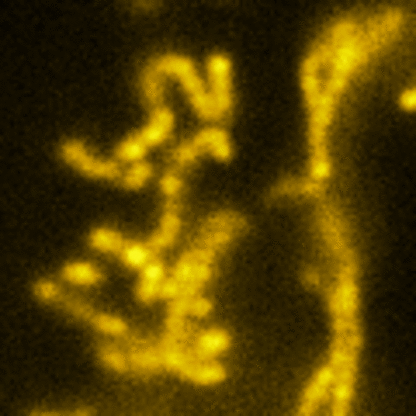
Lattice SIM
Three-dimensional point patterns, referred to as Lattice SIM, for SIM have been described previously by [Betzig] and [Heintzmann]. This is the technology used by the ELYRA 7. In Lattice SIM, the spot pattern is shifted laterally, without rotation steps, which leads to faster imaging and allows live cell imaging of dynamic processes. The image bellow shows simulations of Linear, Quadratic Lattice and Hexagonal Lattice light patterns. The Elyra 7 uses a quadratic lattice pattern for structured illumination microscopy.
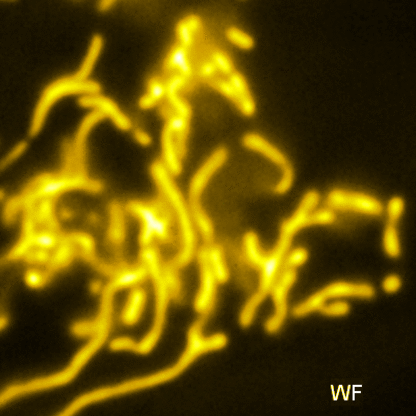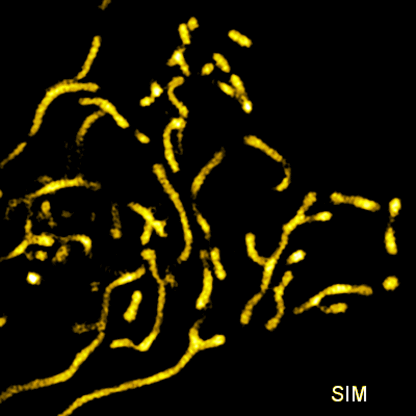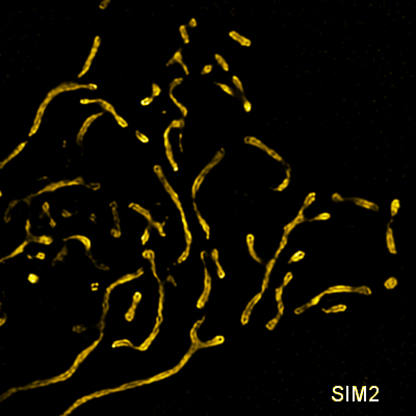
Related articles
Single Molecule Localization Microscopy (SMLM) is the common name of several techniques, which utilizes the blinking of fluorophores to surpass the diffraction limit, such as PALM, dSTORM etc. In all of these techniques, a small subset of fluorophores are imaged in each frame of a long time series. The localization of each detected flourophore is found and the final superresolution image is reconstructed from all localization positions.